The Bioinformatics Core no longer manages the Ingenuity Pathway Tools Site License. Labs that wish to access this program can contact Ingenuity directly (Susan DiIorio; sdiiorio@ingenuity.com).
Below we have listed a number of freely available pathways tools which might meet your analysis needs. We do not have any direct experience with any of these and can only offer limited support in this area.
GeneMANIA (http://www.genemania.org/)
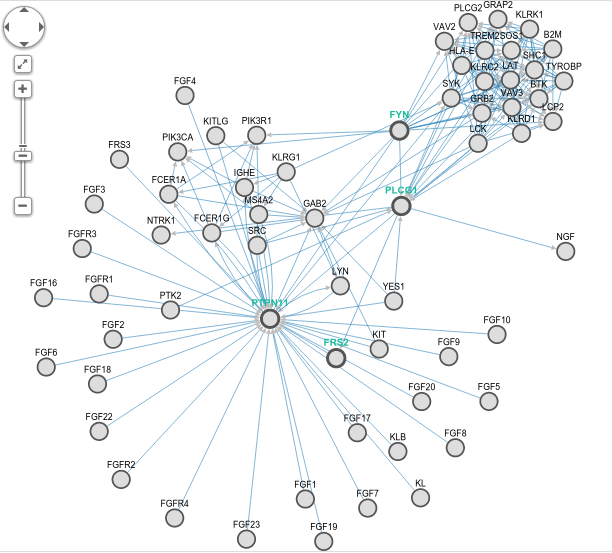
GeneMANIA starts with a set of genes of interest, and brings a network with other genes that are functionally related. For instance assume you have 4 genes that came from results of an experiment, FYN, PLCG1, FRS2, and PTPN11. GeneMANIA generates the below network for you.
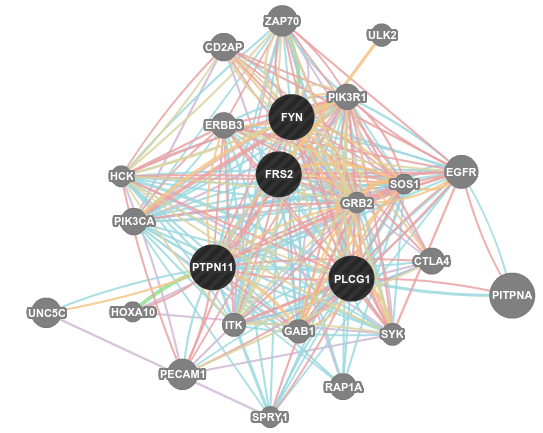
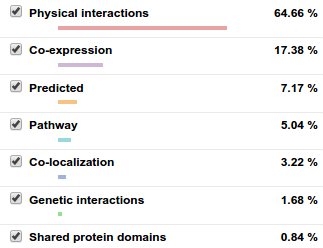
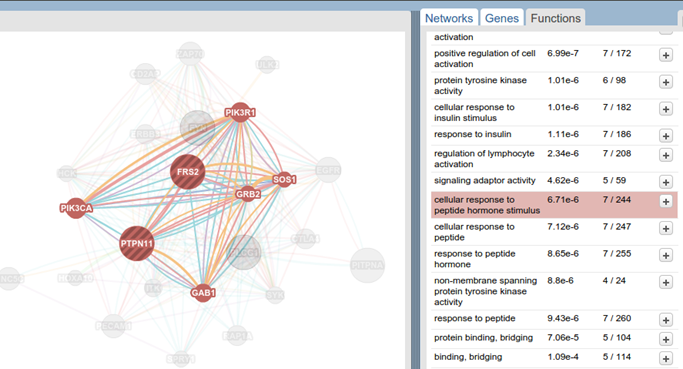
GeneMANIA uses a PageRank-like metric (the one Google uses) to find related genes of your query. At the right, you can see the relation types that GeneMAINA uses. You can always focus on specific relation types of your interest. It has also very cool features like listing the related functions and highlighting the sub-graphs that is related to a selected function (see below image).
PCViz (http://www.pathwaycommons.org/pcviz/)
This is a web front-end to the database Pathway Commons, which integrates publicly available human pathway data. Pathway Commons currently have data from Reactome, NCI Pathway Interaction Database, PANTHER Pathways, PhosphoSitePlus, HumanCyc, and HPRD, and this is growing. Below is the PCViz result for the same 4 genes of interest. In this graph, edges are directed and they mean the source molecule controls-state-change-of the target molecule.
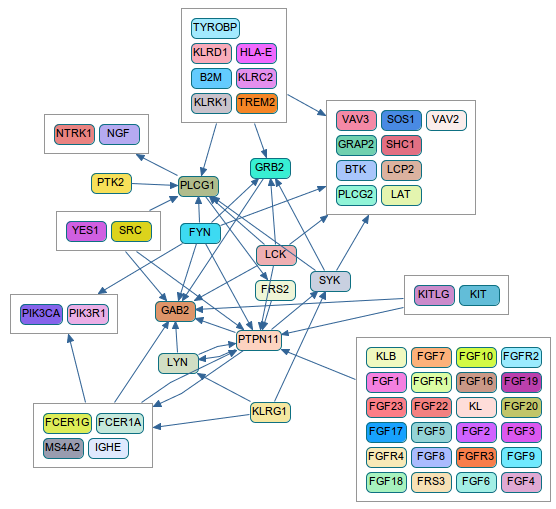
ChiBE (http://code.google.com/p/chibe)
ChiBE is anohther front-end for Pathway Commons. This is not web-based, but a desktop application with many different querying, visualization, and analysis options for pathways. Below graph is the ChiBE version of the above graph. Here, molecules are grouped if their connections with the surrounding genes are similar, for a less complex view.
ChiBE can also show details of these relations. For instance above graph says that FCER1G controls-state-change-of GAB2, and you want to know what that really is. Then, all you need to is to click on that edge and go to the details (below).
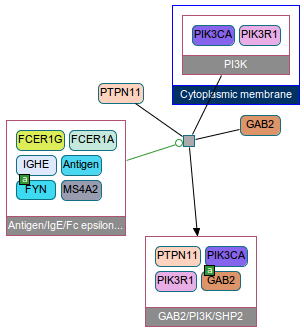
Here you see that FCER1G is part of an antiogen-IG complex, and can activate the recruit of GAB2 to the PI3K complex in the cell membrane.
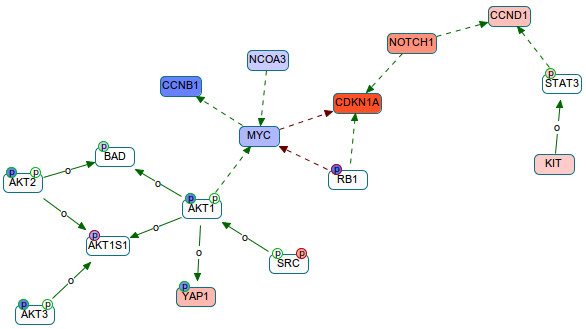
If you want to analyze proteomics data, we are currently developing a fancy feature into ChiBE that highlights the possible cause-effect relations between the measurements. For instance the below graph shows such relations when the cancer drug JQ1 is administered to OVCAR4 cell line (RPPA experiment). Here, p-boxes indicate phosphorylation sites, and the colors indicate changes in measurements. Solid edges are phosphorylation, and dashed edges are expression relations. Background color of the gene is used for the total protein change, and background of p-boxes is used for phospho-changes. This says that the reduction in phospho-AKT1S1 can be due to the reduction in phospho-AKT.